SpaCy IRL 2019 - Wikidata-based NER in v3.0
Published:
On July, 6th in Berlin I attended spaCy IRL - a conference organized by explosion.ai and spacy which you probably know as one of the most popular, powerful and fast NLP libraries. Here is a short overview of the event.
We use spaCy in our daily work as well, so we couldn’t miss the chance to meet the community and exchange research ideas as well as thoughts and expectations on the future of spaCy. Keynotes were not only covering applications of spaCy in research and industry but also more broad and diverse topics. There exist already great highlights (like this and that), so in this post I’d like to go more into details of particular talks.
Wikidata-based NER to appear in spaCy 3.0

Sofie Van Landeghem delivered a great talk (here are the slides) on the upcoming changes in the Named Entity Recognition (NER) module in the next major spaCy version.
In the current version, basic NER is fixed to a predefined set of entity types which you can manually extend. Nevertheless, the prediction quality even in larger models was not perfect so community wanted to have something more robust and customizable for domain-specific tasks.
Uniquely identified named entities? And with synonyms? And formally described? Sounds like a knowledge graph, right? For instance, Wikidata? ![]()
SpaCy think the same! So they took the latest version of Wikidata, sliced it from 50M entities to 1M based on the amount of incoming edges (interwiki links) retaining about 1.5M aliases, i.e., synonyms of named entities. The training process is as follows:
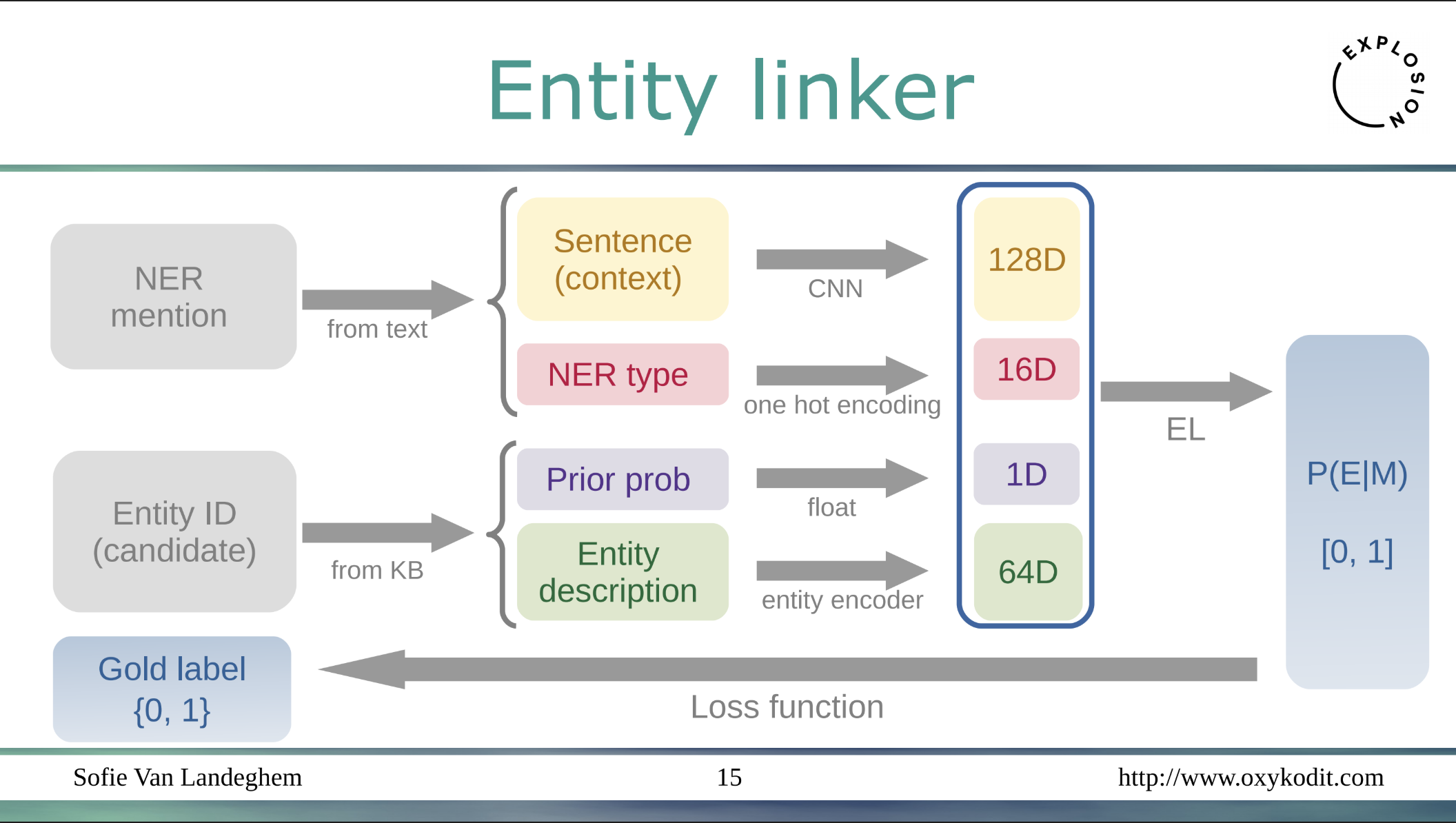
Wikitext with named entities mentions is passed through a context encoder to obtain a 128D vector, 16 supported entity types comprise a 16D vector, then from the extracted Wikidata they encode entity description into a 64D vector, and finally concatenate the vectors together with the 1D prior probability vector. And the classifier is trained to maximize the probability of a Wikidata entity ID, entity type for a given piece of text. What is the accuracy of this approach?
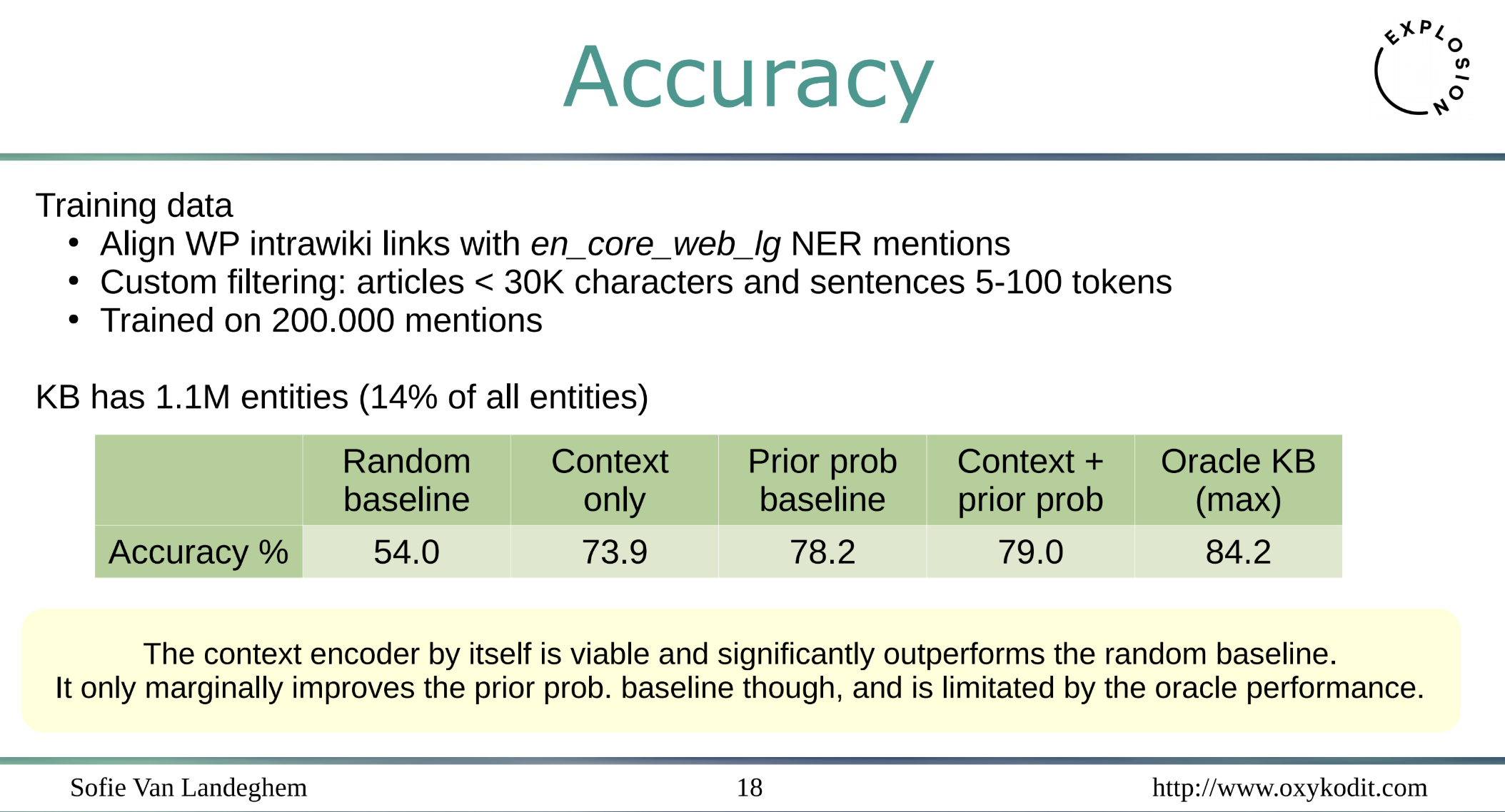
Well, it achieves decent 80% with 200K mentions on 1.1M entites - a nice initial result, and the trained model is just about 350MB. What is even more fascinating - you don’t need huge amounts of text to train a NER component as the following slide shows:
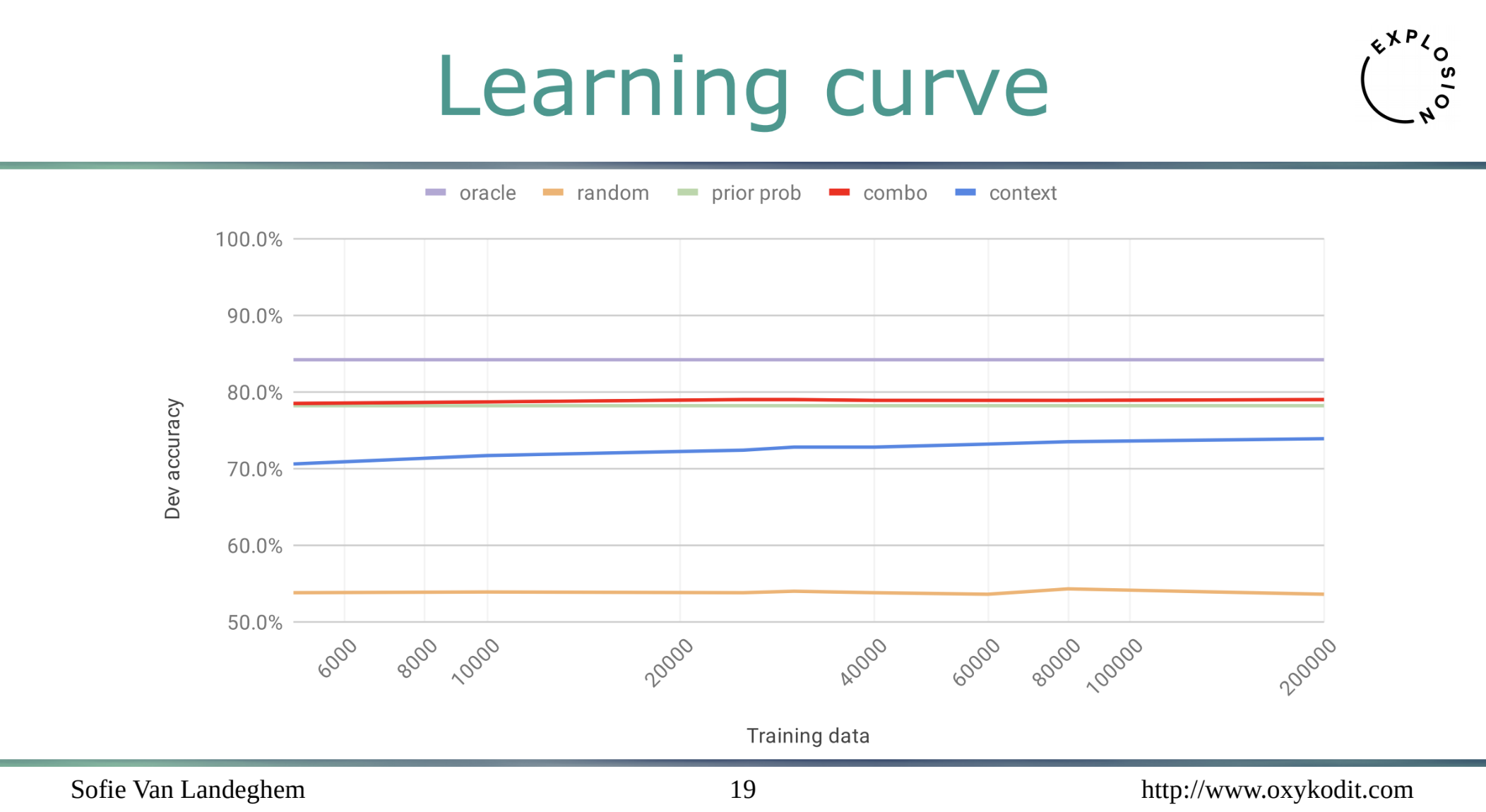
It’s actually fine to have just 6-10K of mentions to achieve almost peak quality! Alright, let me repeat it in simpler words: the approach allows to plug in your own knowledge graph with domain-specific entities and train a NER system with relatively small amounts of data, that’s awesome ![]() And the following talk gave an application example in the biomedical domain.
And the following talk gave an application example in the biomedical domain.
scispaCy

Mark Neumann from Allen AI presented scispaCy, a spaCy-based package for processing biomedial, clinical or scientific texts (slides). Open domain general purpose NER systems have little coverage of biomedical entities. They can probably identify DNA as a named entity, but struggle to link something as complex as “17beta-estradiol”. So the team took two major steps:
- trained a NER system on UMLS, the largest biomedical vocabulary
- employed small amount of rules to identify abbreviations, e.g., phosphatidylinositol-3-kinase (PI3K)
Given just that, they already achieve quite impressive results:
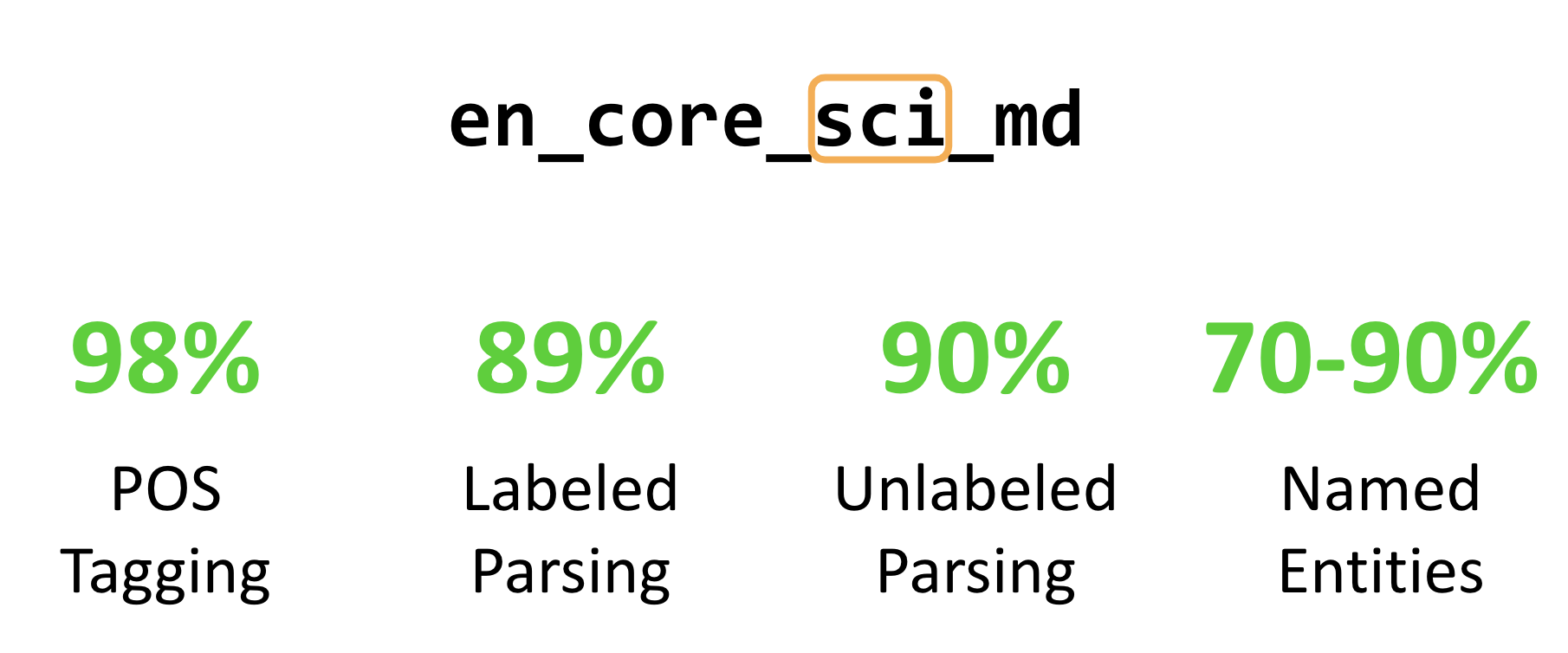
Well, it you were looking how to adapt NER systems to your domain, say, finance, law, or hydrodynamics, looks like we have a working solution now - take (or build) a rich knowledge graph of your domain (and KGs are inherently cool, too), get some decent volumes of training data (come on, 6-10K is not a leviathan after all), put them into spaCy v3.0 and you’re done ![]()
Conclusions
Video recordings are now available as a playlist on YouTube and I encourage you to check out all of them - thanks to the speakers for informative but not-rocket-science-complex talks! More photos are available in the gallery on the official website, credit goes to Ema Discordant. Thanks spaCy for bringing together the audience from so many fields with diverse backgrounds, e.g., I was not the only KG guy in the room :)
See you next time!


